An example of a Distruct chart from a Structure analysis:
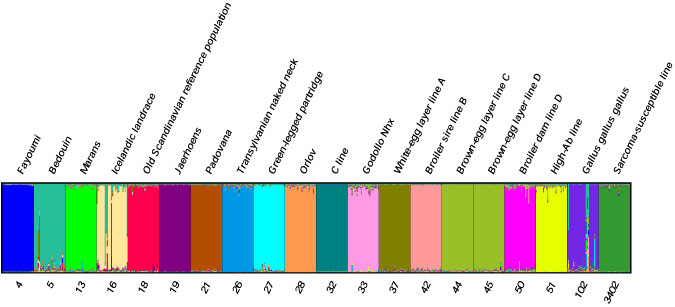 Distruct requires two bits of information from your Structure output file '*_run_1_f' found in the 'Results' directory of your
Structure project. You will edit the following 5 files for each Distruct chart:
Distruct requires two bits of information from your Structure output file '*_run_1_f' found in the 'Results' directory of your
Structure project. You will edit the following 5 files for each Distruct chart:- *.popq (input file of population Q's)
- *.indivq (input file of individual Q's)
- *.names (input file of labels for figure)
- *.perm (colors assigned to pops)
- drawparms (parameters file)
$cp casia.popq trach.popq
If you are running multiple analyses for different k values, I recommend making unique *.popq etc. files for each run. For e.g., trach2.popq, trach3.popq, trach4.popq... This will make for a quick and easy way to run Distruct for all Structure analyses. The 'drawparam' file is the only file read by Distruct and it needs to be edited each time you run Distruct. There are many adjustable parameters in the 'drawparms' file. But I will only cover the minimum edits required. The following is the beginning of the 'drawparms' file. Highlighted in dark grey are the parameters that should be edited.PARAMETERS FOR THE PROGRAM distruct. YOU WILL NEED TO SET THESE IN ORDER TO RUN THE PROGRAM. "(int)" means that this takes an integer value. "(B)" means that this variable is Boolean (1 for True, and 0 for False) "(str)" means that this is a string (but not enclosed in quotes) "(d)" means that this is a double (a real number). Data settings #define INFILE_POPQ trach8.popq // (str) input file of population q's #define INFILE_INDIVQ trach8.indivq // (str) input file of individual q's #define INFILE_LABEL_BELOW trach.names // (str) input file of labels for below figure #define INFILE_LABEL_ATOP trach.names // (str) input file of labels for atop figure #define INFILE_CLUST_PERM trach8.perm // (str) input file of permutation of clusters to print #define OUTFILE Trach_k8.ps //(str) name of output file #define K 8 // (int) number of clusters #define NUMPOPS 5 // (int) number of pre-defined populations #define NUMINDS 43 // (int) number of individuals
Two of the five listed files require data to be copied from the Structure output file (*_run_1_f). For the *.popq file, copy and paste the dark grey highlighted matrix (shown below) from the '*_run_1_f' file into the *.popq file.-------------------------------------------- Proportion of membership of each pre-defined population in each of the 10 clusters Given Inferred Clusters Number of Pop 1 2 3 4 5 6 7 8 9 10 Individuals 1: 0.001 0.001 0.001 0.000 0.001 0.001 0.000 0.996 0.000 0.000 17 2: 0.011 0.034 0.008 0.559 0.034 0.081 0.053 0.067 0.049 0.103 24 3: 0.029 0.020 0.715 0.111 0.042 0.018 0.036 0.000 0.023 0.007 15 4: 0.487 0.010 0.248 0.104 0.023 0.079 0.041 0.000 0.007 0.002 14 5: 0.070 0.005 0.172 0.273 0.008 0.025 0.426 0.000 0.015 0.006 13 --------------------------------------------
For the *.indivq file, copy and paste the dark grey highlighted matrix (shown below) from the '*_run_1_f' file into the *.indivq file. Note that the order of the individuals in this file dictate the order in the Distruct chart.Inferred ancestry of individuals: Label (%Miss) Pop: Inferred clusters 1 gaig_AHP116 (75) 1 : 0.000 0.000 0.000 0.000 0.000 0.000 0.000 1.000 0.000 0.000 2 gaig_AHP280 (78) 1 : 0.002 0.003 0.003 0.001 0.002 0.003 0.002 0.983 0.001 0.001 3 gaig_AHP280 (82) 1 : 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.999 0.000 0.000 4 gaig_TP2943 (67) 1 : 0.001 0.001 0.001 0.000 0.001 0.001 0.001 0.992 0.001 0.001 5 gaig_TP2991 (65) 1 : 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.999 0.000 0.000 6 gaig_TP2991 (84) 1 : 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.999 0.000 0.000 7 gaig_TP3004 (81) 1 : 0.000 0.000 0.000 0.000 0.000 0.000 0.000 0.999 0.000 0.000 8 e_AHP2715 (92) 2 : 0.004 0.014 0.005 0.380 0.008 0.021 0.029 0.000 0.006 0.532 9 e_AHP2722 (78) 2 : 0.017 0.046 0.002 0.507 0.112 0.175 0.082 0.000 0.014 0.045 10 e_AHP2731 (79) 2 : 0.019 0.004 0.004 0.314 0.031 0.568 0.009 0.000 0.050 0.001 11 e_AHP2746 (89) 2 : 0.031 0.042 0.002 0.504 0.038 0.353 0.008 0.000 0.011 0.011 12 e_AHP2759 (89) 2 : 0.002 0.054 0.004 0.808 0.007 0.067 0.032 0.006 0.017 0.003 13 e_AHP2765 (78) 2 : 0.003 0.036 0.005 0.681 0.106 0.069 0.012 0.081 0.005 0.001 14 e_JFP644 (85) 2 : 0.012 0.034 0.002 0.293 0.010 0.014 0.329 0.000 0.293 0.013 15 e_MVZ250717 (90) 2 : 0.007 0.003 0.001 0.793 0.037 0.008 0.025 0.021 0.061 0.044 16 e_TP29441 (78) 2 : 0.005 0.002 0.001 0.473 0.002 0.075 0.012 0.247 0.012 0.171 17 e_TP29442 (82) 2 : 0.005 0.067 0.001 0.637 0.015 0.034 0.014 0.222 0.001 0.003 18 e_TP29700 (74) 2 : 0.004 0.005 0.003 0.266 0.072 0.209 0.004 0.000 0.415 0.023 19 e_TP29715 (78) 2 : 0.003 0.009 0.002 0.593 0.013 0.063 0.003 0.000 0.028 0.286 20 e_TP29950 (99) 2 : 0.047 0.060 0.052 0.548 0.056 0.053 0.056 0.004 0.062 0.062 21 e_TP29957 (98) 2 : 0.025 0.038 0.020 0.643 0.059 0.040 0.028 0.005 0.044 0.098 22 e_TP29959 (98) 2 : 0.015 0.043 0.016 0.589 0.054 0.036 0.026 0.133 0.018 0.072 23 e_TP29969 (92) 2 : 0.008 0.008 0.008 0.662 0.015 0.023 0.005 0.247 0.008 0.016 24 e_TP29971 (99) 2 : 0.049 0.055 0.059 0.475 0.057 0.050 0.050 0.080 0.060 0.064 25 e_TP30028 (70) 2 : 0.003 0.151 0.002 0.752 0.026 0.004 0.015 0.025 0.008 0.013 26 e_TP30030 (77) 2 : 0.001 0.076 0.001 0.810 0.076 0.003 0.024 0.000 0.001 0.007 27 e_TP30036 (81) 2 : 0.004 0.026 0.001 0.577 0.003 0.055 0.002 0.081 0.006 0.244 28 e_TP30046 (76) 2 : 0.003 0.014 0.001 0.727 0.006 0.002 0.001 0.228 0.011 0.006 29 e_TP30059b (89) 2 : 0.004 0.014 0.005 0.696 0.007 0.024 0.006 0.227 0.013 0.005 30 e_TP30070 (84) 2 : 0.001 0.002 0.001 0.389 0.006 0.006 0.485 0.000 0.004 0.106 31 e_TP30072-2 (77) 2 : 0.002 0.007 0.001 0.309 0.004 0.006 0.007 0.000 0.023 0.641 32 ExS_N74672 (85) 3 : 0.109 0.004 0.487 0.149 0.002 0.031 0.144 0.000 0.071 0.003 33 ExS_N74673 (91) 3 : 0.018 0.011 0.763 0.119 0.050 0.019 0.007 0.000 0.005 0.007 34 ExS_N74674 (68) 3 : 0.003 0.074 0.630 0.084 0.122 0.036 0.020 0.000 0.022 0.008 35 ExS_N74675 (87) 3 : 0.004 0.004 0.802 0.145 0.022 0.004 0.003 0.000 0.007 0.009 36 ExS_TP29746 (86) 3 : 0.009 0.007 0.893 0.058 0.013 0.002 0.004 0.000 0.009 0.006 37 tss_TP28037 (82) 4 : 0.165 0.020 0.002 0.258 0.075 0.307 0.160 0.000 0.010 0.003 38 tss_TP28323 (74) 4 : 0.858 0.007 0.009 0.109 0.003 0.003 0.001 0.000 0.009 0.001 39 tss_TP28331 (70) 4 : 0.919 0.008 0.003 0.048 0.011 0.006 0.001 0.000 0.003 0.002 40 tss_TP29723 (57) 4 : 0.005 0.004 0.980 0.000 0.003 0.001 0.001 0.000 0.005 0.001 41 tst_N74447 (90) 5 : 0.125 0.005 0.452 0.259 0.006 0.008 0.107 0.000 0.034 0.004 42 tst_TP30111 (88) 5 : 0.084 0.003 0.031 0.334 0.012 0.062 0.458 0.000 0.008 0.008 43 tst_TP30114 (67) 5 : 0.001 0.006 0.034 0.228 0.005 0.003 0.714 0.000 0.003 0.005
The '*.names' file is where you tell Distruct the names of the populations1 Gaig 2 TSE 3 TSExTSS 4 TSS 5 TST
The '*.perm' file (I named mine trach.colors) defines colors on the chart (refer to Distruct manual for color options and themes):1 orange 2 blue 3 pink 4 green 5 red 6 purple 7 light_green 8 light_yellow
You are now ready to create a chart with Distruct 1.1$ cd /pathto/distruct1.1 $ ./distructLinux1.1
I recommend NOT opening the *.ps output file on the Ubuntu server. I prefer to send it to my PC and edit it in Illustrator.